Hi! We're
Matt and John
We're creative technologists probably best known for our crypto art projects the CryptoPunks and Autoglyphs.
MORE ABOUT US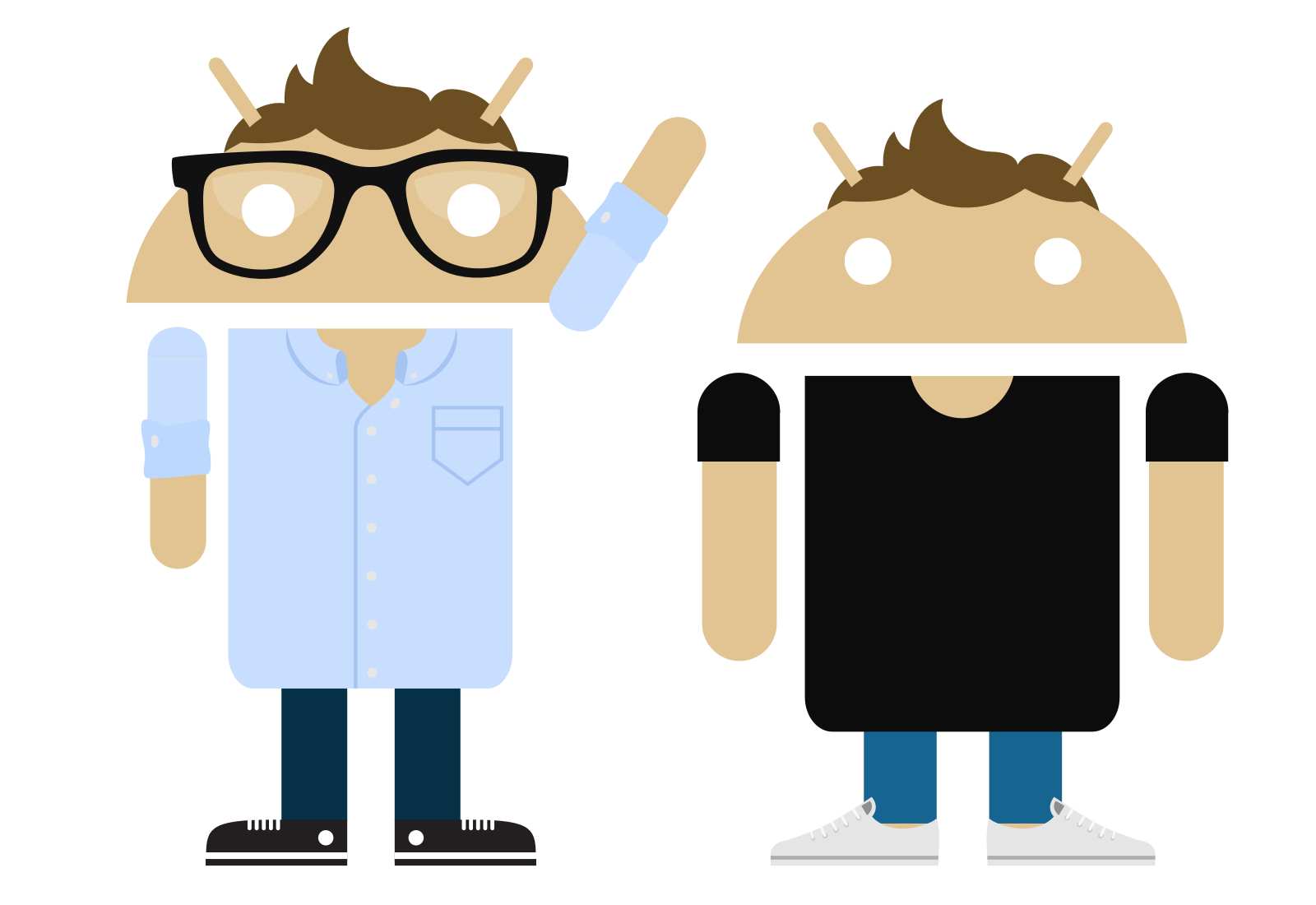
The Cryptopunks
10,000 unique collectible characters with proof of ownership stored on the Ethereum blockchain.
READ MORE
Data Visualizations for Flutter Web
An interactive data visualization created for Google to launch Flutter Web at I/O 2019.
READ MORE
Androidify
with Google
A small internal project with Google that grew to become the face of the Android global brand.
READ MORE